Key takeaways
-
Researchers sought to understand how low- and high-grade gliomas develop resistance to BRAF/MEK inhibitors.
-
They identified several potential mutations plus a subset of patients without a clearly defined resistance mechanism.
-
This will allow physicians to tailor relapse therapy for each patient to get the best clinical response.
-
Results will also help identify other potential combination therapies for future clinical trials.
Background: Gliomas develop resistance to RAF and MEK inhibitors
Extracellular signal-regulated kinase (ERK) signaling pathway mutations are common in low-grade gliomas (LGG) and high-grade gliomas (HGG). The BRAFV600E mutation is common and causes the BRAF protein signal cells to uncontrollably grow independent of upstream regulators.
Selective mutant RAF inhibitors (RAFi) have, to varying degrees, successfully targeted the BRAFV600E mutation in colon, lung, melanoma, and other cancers. Treatment-emergent resistance in melanoma prompted the use of RAFi and MEK inhibitor (MEKi) combinations to improve response. Early results demonstrated response in using the combination in some gliomas, especially LGG, but many tumors do not respond or respond only briefly.
Known RAFi resistance mechanisms include:
- Aberrantly spliced BRAF
- Mutations in RAS (a common oncogene)
- Loss of neurofibromatosis type 1 (NF1)
- RAF proto-oncogene serine/threonine-protein kinase (CRAF) switching
- Increased human epidermal growth factor receptor (EGFR) kinase activation
It is unknown whether gliomas develop resistance using one or more of these mechanisms or via a unique, lineage-specific mechanism. To effectively use molecularly targeted therapies in patients with gliomas, it is critical to understand how LGGs and HGGs develop resistance to AF inhibitor/MEK inhibitor. One research barrier is the scarcity of pre- and post-treatment specimens.
As part of a multi-institutional collaboration, neuro-oncology researchers, including Jean Mulcahy-Levy, MD, from Children’s Hospital Colorado who belong to the Morgan Adams Foundation Pediatric Brain Tumor Research Program identified resistance mechanisms, including a unique RAFi resistance mechanism that promotes BRAF dimerization to evade cell growth inhibition. In this study, researchers investigated and identified additional drug resistance mechanisms.
Methods: pre- and post-treatment LGG and HGG samples prepared and sequenced
Researchers obtained paired tissue samples for 15 patients with LGG or HGG before and after treatment with RAFi or RAFi/ MEKi and queried them for mechanisms of resistance in BRAFV600E glioma models.
Comparing pre- and post-tissue sample genomic sequences
- Cells plated, treated and monitored for growth
- Ribonucleic acid (RNA) extracted from paired samples
- Next-generation sequencing (NGS) performed on pre- and post-RAFi treated tissue to identify potential genomic resistance mechanisms
- Removed shared somatic variants
- Two cell lines derived from patients with BRAFV600E-mutant glioma evaluated for treatment-emergent resistance
- Stable NF1 knockdown lines created using two different Clustered Regularly Interspaced Palindromic Repeats single-guide RNA constructs
- CBL (E3 ubiquitin-protein ligase) knockdowns tested for sensitivity to RAFi (vemurafenib), MEKi (cobimetinib), and EGFRi (neratinib), alone or in combination
- Stably expressed phosphate and tensin homolog (PTEN) in B76 cells using retroviral infection with puromycin evaluated role of PTEN loss in B76 sensitivity
- RNA sequencing (RNA-seq) performed on 12 sample pairs
- RAF1 knockdowns to test sensitivity to RAFi
- Effect of treatment with BRAFV600E inhibitor, vemurafenib, pan-RAF inhibitor LY3009120, or combination tested on ERK activity
- Bulk RNA-seq performed on nine paired patient samples and three patient-derived cell line RAFi-sensitive/resistant pairs
Results: several potential mechanisms of resistance to BRAFi identified
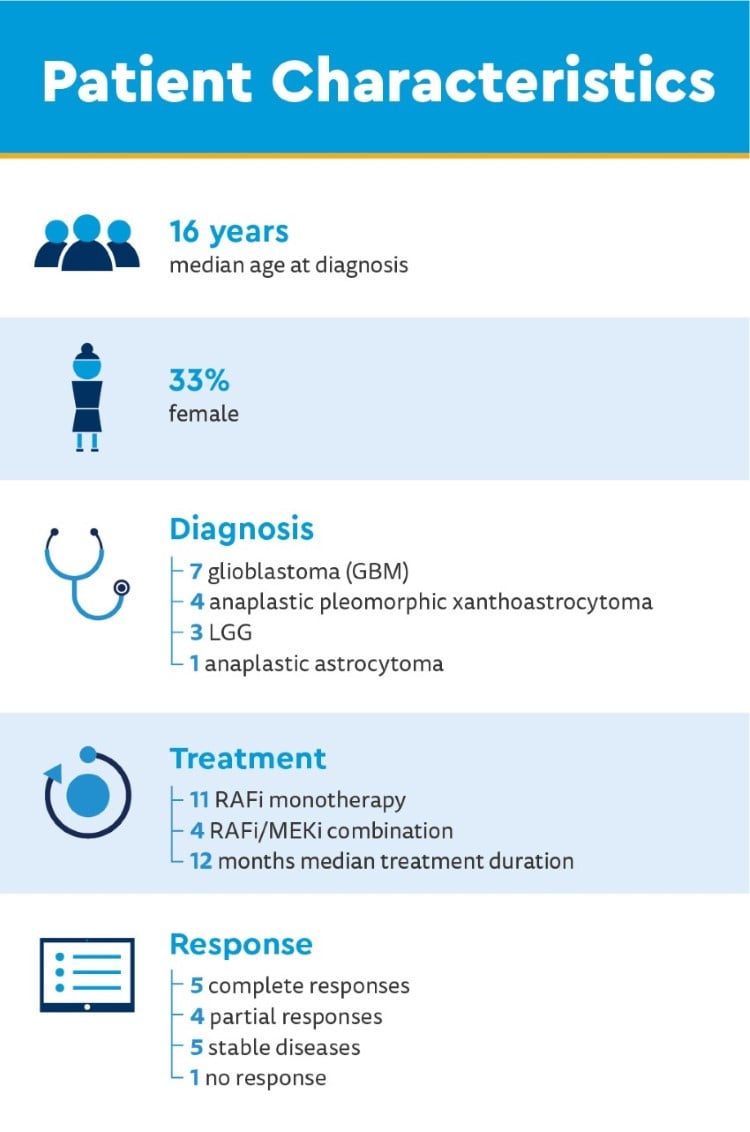
Clinical genomic sequencing of pre-treatment tissue findings
- 80% homozygous loss of CDKN2A/B
- Common co-occurrence in BRAF alterations
- Associated with worse prognosis
- 20% TERT promoter mutations or gene amplifications
- 13% missense mutations in TP53
- 7% missense mutations in ATRX
Researchers assembled 12 of the most frequently mutated genes in glioblastoma, but no additional mutations genes were found.

New post-treatment mutations and tumor progression
Analysis of 15 tissue sample pairs identified 13 alterations conferring putative resistance among nine paired samples (including mutations involving ERRFI1, BAP1, ANKHD1 and MAP2K1).
- 60% of patient-derived paired tumor samples had presumed etiologies for RAFi resistance
- All present in subclonal variant allele frequencies (VAF)
- Alterations identified in each gene linked with BRAF alterations
- 40% of paired samples had no alteration identified
- Possible adaptive resistance instead of genomic
- BRAF-mutant glioma escapes targeted therapy along conserved pathways expressed in a subset of GBM
Alterations found in vivo
- ERRFI1S251 (loss-of-function mutation) and TET2V1199E (epigenetic DNA modification) observed in resistant clones as possible mechanisms of resistance in vitro
- NF1 alterations decreased glioma sensitivity to RAFi, partial resistance with RAFi/MEKi
- Loss of CBL conferred relative resistance to single drug and combination therapy, sensitivity to RAFi/MEKi or RAFiþ/GFRi relatively good
- Restoration of PTEN mildly increased sensitivity of B76 to vemurafenib
- BRAFV600E inhibitor, vemurafenib, pan-RAF inhibitor LY3009120 and combination inhibited ERK activity
- CRAF inhibitor, ZM336372, alone had little effect on RAFi; significant inhibition when combined with vemurafenib
Discussion: therapeutic impact of understanding RAFi resistance
The role of BRAFV600E mutations in glioma pathogenesis and prognosis is well described in pediatric glioma and increasingly studied in adult glioma. Multiple case reports and early-phase clinical trials support RAF-targeted therapy in patients with BRAFV600E alterations, though resistance is common.
Understanding RAFi resistance is key to developing targeted therapies. This study identified and validated a range of genomic and adaptive mechanisms of resistance.
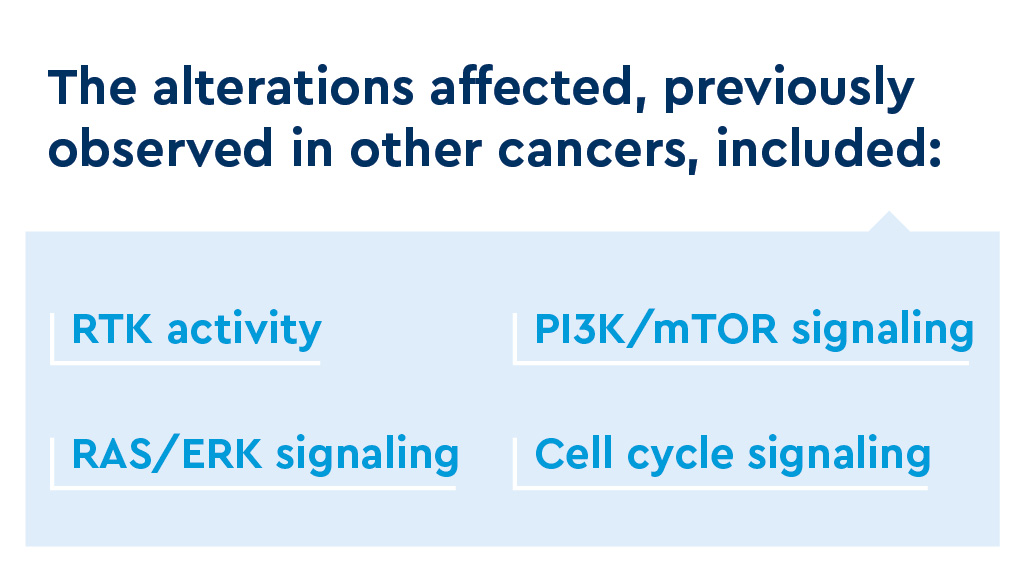
Key study findings:
- Small genetic alterations may be sufficient for clinical resistance
- Loss of tumor suppressor (NF1 and PTEN) can confer relative resistance to targeted therapies
- Two primary patterns of adaptive resistance to RAFi found, suggesting adaptive resistance emerges along conserved pathways
- Driven by increased TGFB1, expression pattern most consistent with mesenchymal glioblastoma subtype
- Indicated by decreased TGFB1, most like proneural glioblastoma subtype
- CRAF upregulation is a mechanism of resistance
- Combined therapy with a dimer-disrupter and MEKi may be effective in patients with BRAFV600E-mutant glioma who progressed on the currently FDA-approved RAFi
- Autophagy is another mechanism of resistance, but its role unclear
This is the largest cohort to date of paired pre-/post-RAFi glioma samples, but due to the rarity of paired tumor samples from patients with glioma, the sample size was a limitation.
Conclusion: BRAFi resistance mechanisms are varied
Resistance mechanisms to BRAFi in glioma are varied and may predict effective precision combinations of targeted therapy. This highlights the importance of a personalized approach and the value of prioritizing tissue collection following progression on targeted therapy.
Featured Researchers

Jean Mulcahy Levy, MD
Pediatric neuro-oncologist
Center for Cancer and Blood Disorders
Children's Hospital Colorado
Associate professor
Pediatric Hematology/Oncology and Bone Marrow Transplantation
University of Colorado School of Medicine





 720-777-0123
720-777-0123